r2aGUI
New: version 2.7.0 now available! Converts to Nifti/Analyze from PAR/REC V4.2 and below
(12/10/2009)
Summary
Programming language: Matlab OS: Linux/Unix/Windows
The R2AGUI converts raw data from PHILIPS MRI scanners (PAR/REC format) to the popular ANALYZE 7.5 format, which is used in SPM (http://www.fil.ion.ucl.ac.uk/spm/, Software package for fMRI post-processing) and many other imaging software packages. Versions from 2.4 and newer also convert to the Nifti format, as used in spm5 and other packages in a unified move to reach file format unity.
IMPORTANT NOTES: we consider the Nifti conversion as experimental, it works for us, but we do not guarantee anything. Check converted files well. USE ONE AND THE SAME R2AGUI VERSION FOR A COMPLETE STUDY! MIXING DATA FROM DIFFERENT VERSIONS IS DANGEROUS! (see Notes below for more info)
R2agui has an easy to use interface, and can batch process many files automatically.
On Philips MR scanners, raw binary image data is stored byte-by-byte in a big *.REC file. When a series of images are made, the *.REC file will contain all the bytes for all images (4D image format). In one *.PAR file (ASCII), all the meta-information such as number of images/slices, voxel size, 8/16/32 bit data format, and many more are stored. R2AGUI reads the PAR file parameters, and correctly converts the REC file to (multiple) 3D Nifti or ANALYZE format images in a separete directory. The ANALYZE format consists of a *.img and *.hdr file per volume, Nifti is one .nii containing raw data and meta information per volume.
R2AGUI can convert many Philips PAR/REC file types, since it correctly interpretes the many PAR file parameters. It can also batch process all PAR/REC files it finds in a directory.
R2AGUI can now handle PAR/REC files created by the Philips Research Tools version 3 (used mostly on Intera type
scanners) as well as ther recent version 4, mostly used on the new Achieva type scanners). In version 4 and 4.2 the
PAR file format changed considerably. It will recognize the Version of the PAR/REC format automatically, and invoke the proper conversion routine.
The interface allows one to select and browse images, see some of the meta information of the selected image in the interface, add a prefix to the filename, and batch process many images.
The software contains a couple of functions that were derived from spm99, which is also relased under the GPL (see http://www.fil.ion.ucl.ac.uk/spm/software/spm99/).
R2AGUI is intended for users with no or little matlab experience, with a research or clinical background. It was developed by researchers from the Utrecht University Medical Center and from the Helmholtz Institute, Utrecht University, the Netherlands (most notably Dr. Bas Neggers and Dr. Hermans).
System Requirements
1. Operating Systems: Linux or Windows (tested on Fedora Core 2 Linux and on Windows XP. Likely to work on most modern Linux distributions and
Windows versions that run matlab, or even Mac OSX).
2. Matlab 6.0 or higher installed (tested on Matlab 6.5 SP1 and 7.0.1 SP1). Does NOT work for matlab 5 and older.
3. Linux: Writing permission in the folder where your PAR/REC files are stored.
Installation instructions
1. Copy the archive (r2agui.tar.gz) to a location on your matlab path. Usually matlab/work is fine, or perhaps your spm directory.
2. Extract the files by typing tar xzvf r2agui.tar.gz (Linux). In Windows, WinZip should be able to extract the 2 files in the archive (r2agui.m and r2agui.fig). Make sure both files are in the same folder, or at least somewhere on the matlab search path.
CAUTION MS-Windows users: WinZip among others are known to not always extract *.tar.gz (as common in Linux) zip files correctly, this could lead to your .fig file in the archive being extracted erroneously which causes r2agui to crash when started. See this link for more information on what to change when unzipping tar.gz format using WinZip.
How to use r2agui
1. Type 'r2agui' at your matlab command prompt, and the GUI should appear (see screenshot below).
2. Click on 'open PAR file' to open a PAR file. Information on the PAR file is displayed.
3. Click on 'Convert' to start conversion. All dynamics in the corresponding REC file will be converted, the analyze files will bes saved in a
separate folder for each PAR/REC file.
4. When enabling the 'Batch Off/On' button, all PAR/REC files within the same folder will be converted in one go.
5. in r2agui_defaults.m you can personalize some settings for your site.
Notes
1. Avoid placing PAR/REC files in folders with spaces in their names. Especially on Windows platforms this seems to lead to problems.
2. You do not have to start spm5/spm2/spm99 to use r2agui versions 2.0 , 2.1, 2.3, 2.4, 2.5(.1) or 2.7, they are standalone tools.
However, in v2.4 and higher spm5 or spm2 file selector GUIs will be invoked when found on your matlab path, because they are
easier to use than some local platform file selectors, especially on *nix systems. SPM(2 or 5) has to be in your path for version 2.2 and below
3. VERY IMPORTANT! Use the same version of r2agui for conversion of all data in your study! Mixing versions can do damage to your results!
4. Use it at your own risk. We are MR researhers working with PAR/REC data and r2agui works fine for us, but we do not guarantee it will work for everyone. We have no formal relationship with Philips Medical Systems nor do we provide any official support for their data.
New in v2.7
Donate for r2agui development
If you wish, you can support the development of r2agui and other open source MR analysis tools by donating money. We will always freely distribute the tools you sponsored with your donation under an open source license, for the benefit of the scientific community.
see also: SPM2-batch, a complete SPM automization engine.
To the files!
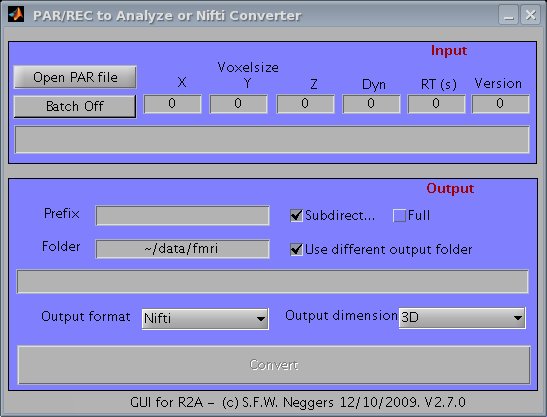
Screenshot of r2agui.